Understanding the EMDB-SFF Data Model¶
Contents
We present a brief synopsis of the data model by outlining the fields and attributes of each entity in the model. You can find the full schema documentation at [LINK]. For each entity we have included a screenshot of the entity’s structure.
The Developing with sfftk-rw guide shows how to create and manipulate corresponding Python objects.
Top Level: An EMDB-SFF Segmentation¶
An EMDB-SFF segmentation is structured as follows:
At the top level, a segmentation has the following attributes.
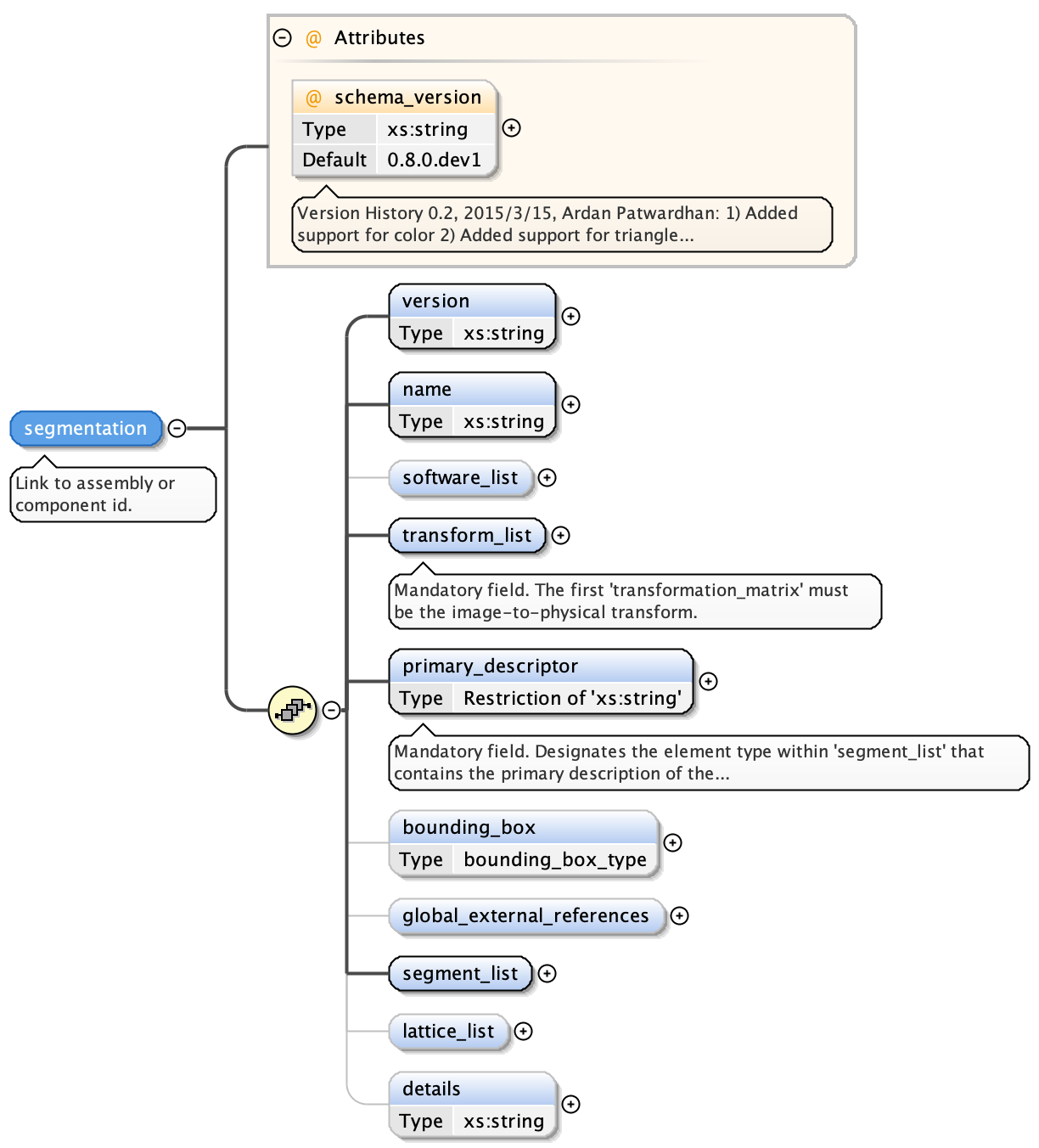
- the version of the EMDB-SFF schema
- the name of the segmentation - free text
- a software_list container (see Software)
- a transform_list container (see Transformation Matrix)
- a primary descriptor field - an indication of the primary geometrical descriptor. The only permitted values are meshList, threeDVolume or shapePrimitiveList.
- the dimensions of the regions bounding box (see Bounding Box)
- a global external references container (see Global External References and External References)
- a segment_list container (see Segments)
- a lattice_list container (see Lattices)
- free text details describing the segmentation
Software¶
Each software item is described by three fields:
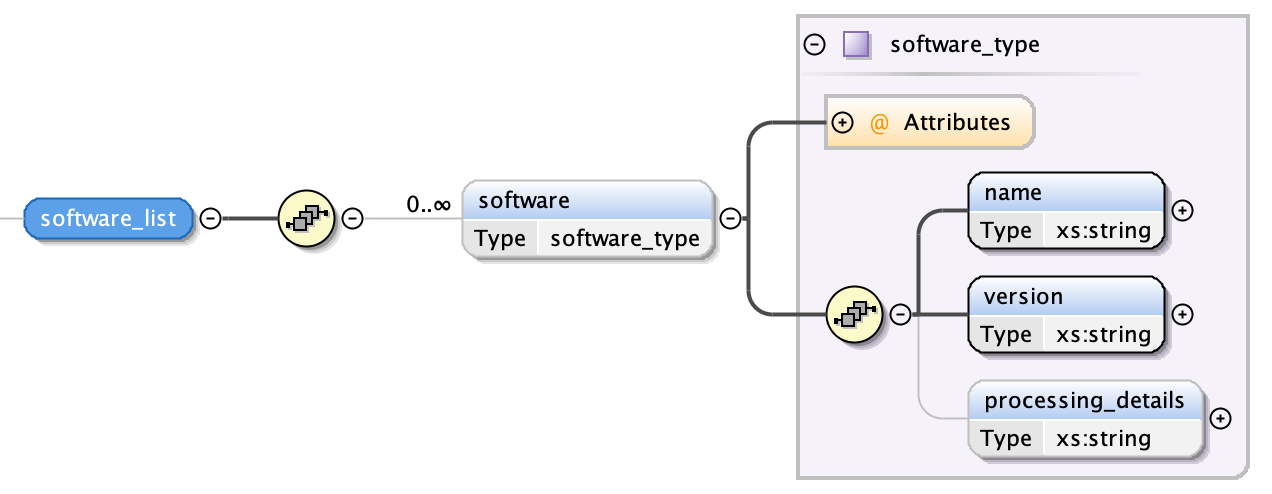
- the name of the software application used to generate the segmentation
- the software’s version
- the processing details by which the segmentation was produced in free text
Transformation Matrix¶
Each transform is a transformation matrix consisting of four fields:
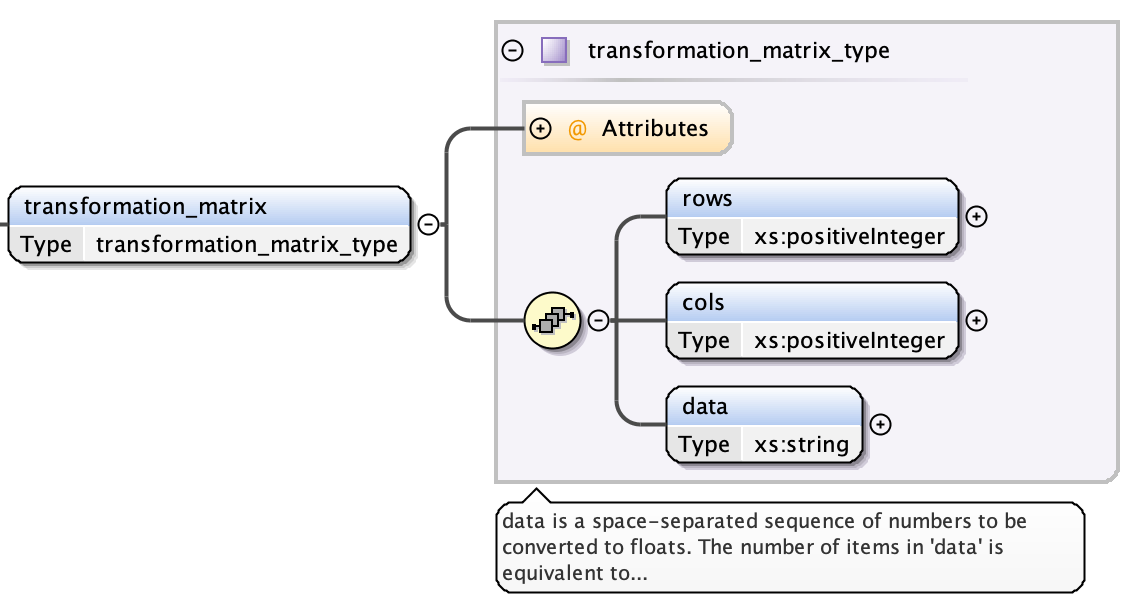
- the transform index attribute - a unique integer value over all transforms
- the number of rows
- the number of columns, and
- a space-separated string of the data in the matrix
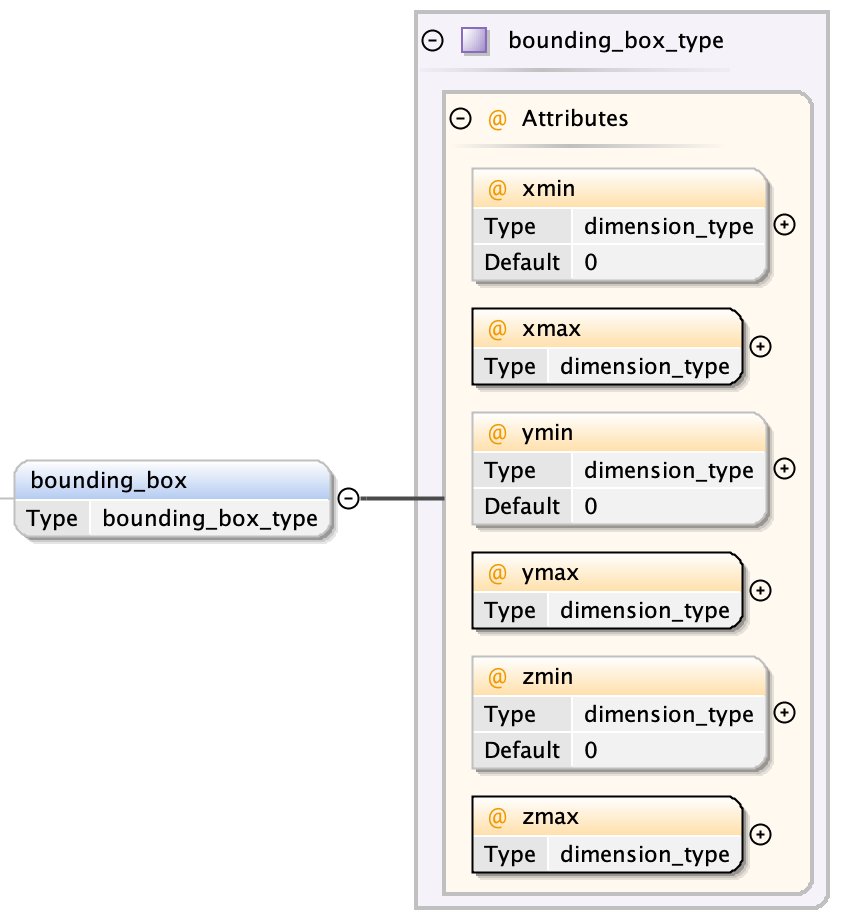
Bounding Box¶
The bounding box consists of minimum and maximum x, y and z values defining the extent of the bounding box (xmin, xmax, ymin, ymax, zmin, zmax)
Global External References and External References¶
A global external reference consists of a set of fixed descriptors by which the overall segmentation may be described. They are distinct from but similar to external references which apply to single segments i.e. a global external reference is to the segmentation while a (local) external reference is to a segment. To make concrete how these are specified, we will use the example of EMPIAR-10070. Suppose we wish to annotate our segmentation (not segment) with this entry. Then the corresponding global external reference will have the following fields.
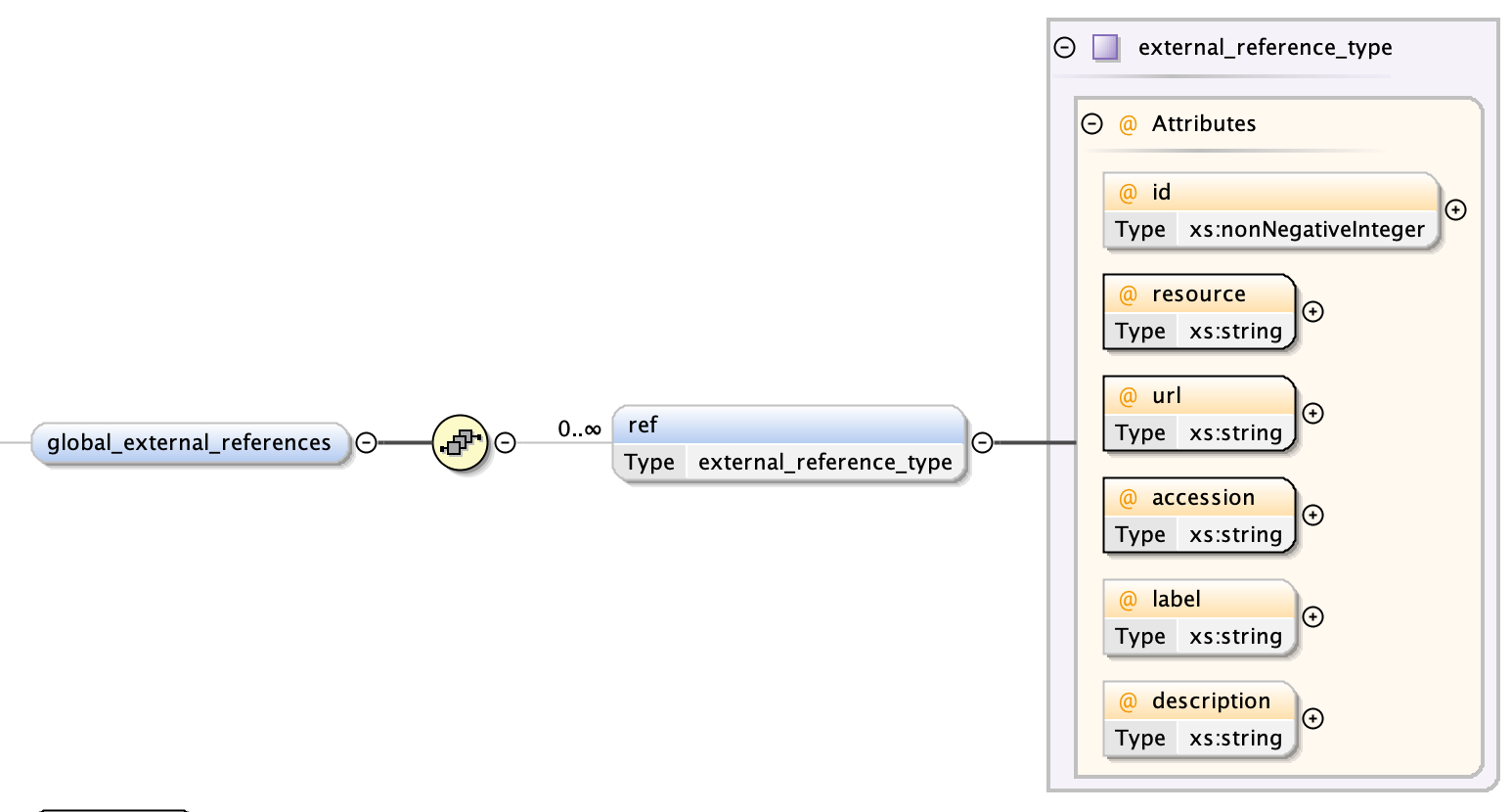
- the type is the name of the resource - from our example this will be “empiar” (case-sensitive). Other examples are “go” for Gene Ontology, “emdb” for EMDB (see Resources for External References for the full list of supported resources)
- the otherType is either a URI or IRI at which the accession (see value) is accessible) - from our example this would be https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10070/
- the value is the complete accession - from our example this is
"10070" - the optional label is a string associated with this entry; for EMPIAR-10070 this is the title of the entry “Focused Ion Beam-Scanning Electron Microscopy of mitochondrial reticulum in murine skeletal muscle”, and
- the optional description is an auxiliary string of the entry; for EMPIAR entries this is the imaging modality “FIB-SEM”
Segments¶
A segment is a complex structure consisting of the following top-level entities:
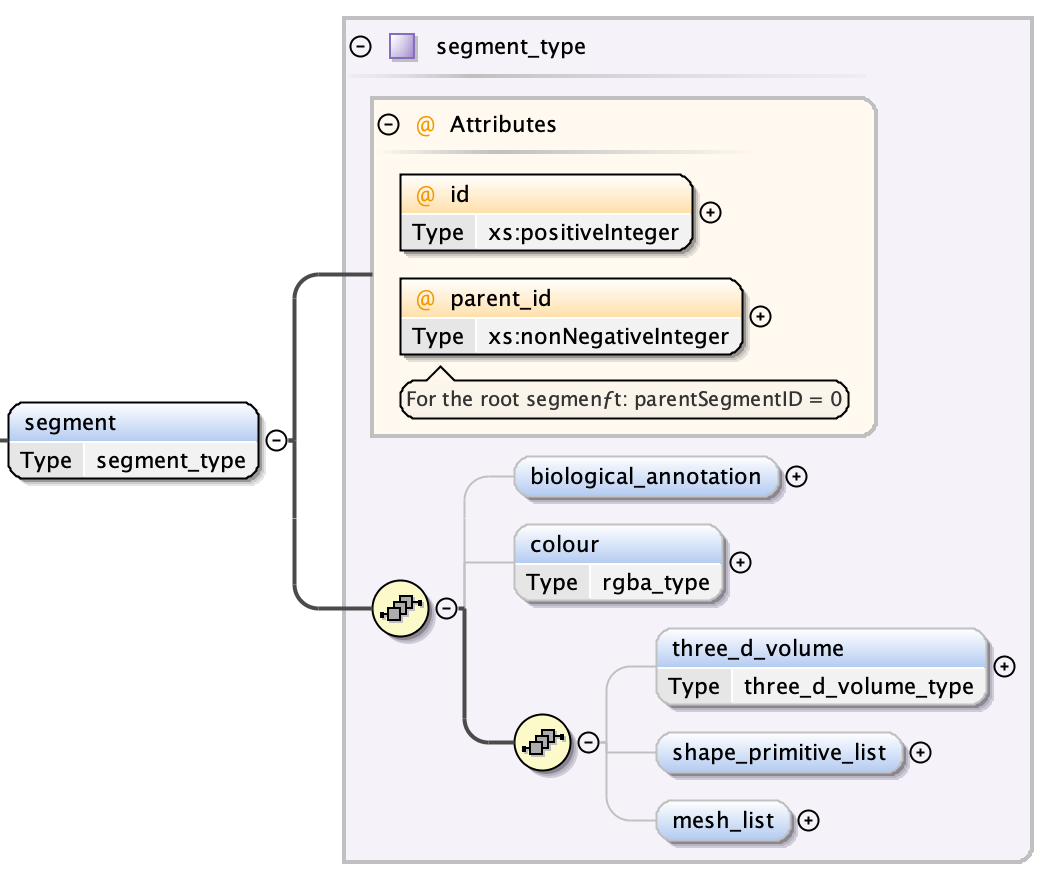
- a biological annotation description (see Segments: Biological Annotation)
- the colour of the segment in arithmetic
rgba - an optional list of meshes (see Segments: Meshes)
- an optional 3D volume description (see Segments: 3D Volumes)
- an optional list of shape primitives (see Segments: Shape Primitives)
Segments: Biological Annotation¶
The biological annotation consists of the following fields:
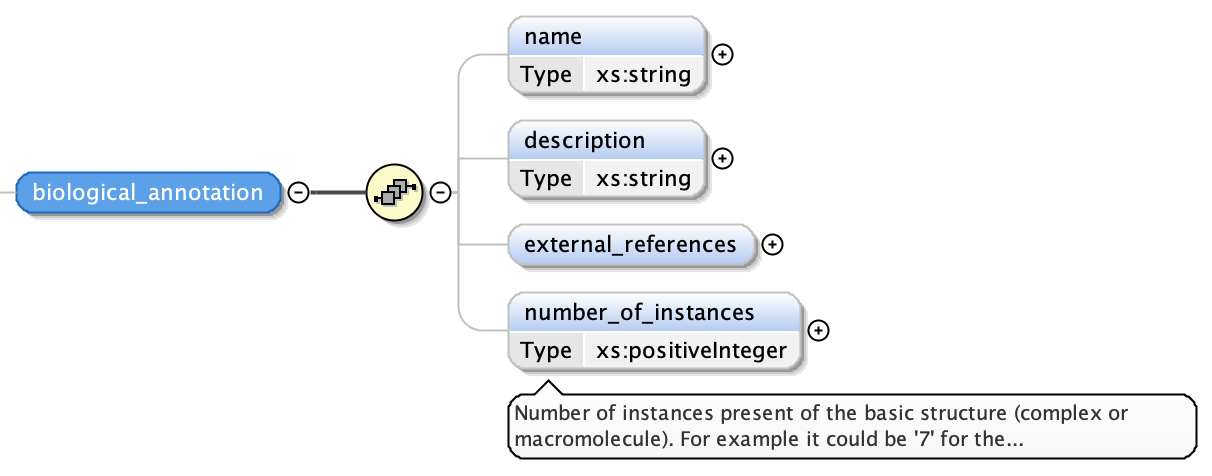
- the name of the segment as a free text string
- a description of the segment as a free text string
- a numerical indication of the number of instances of this segment; this has a default value of one (1)
- a list of external references similar to global external references described in Global External References and External References
Segments: Meshes¶
A mesh has three fields, each of which are further structured:
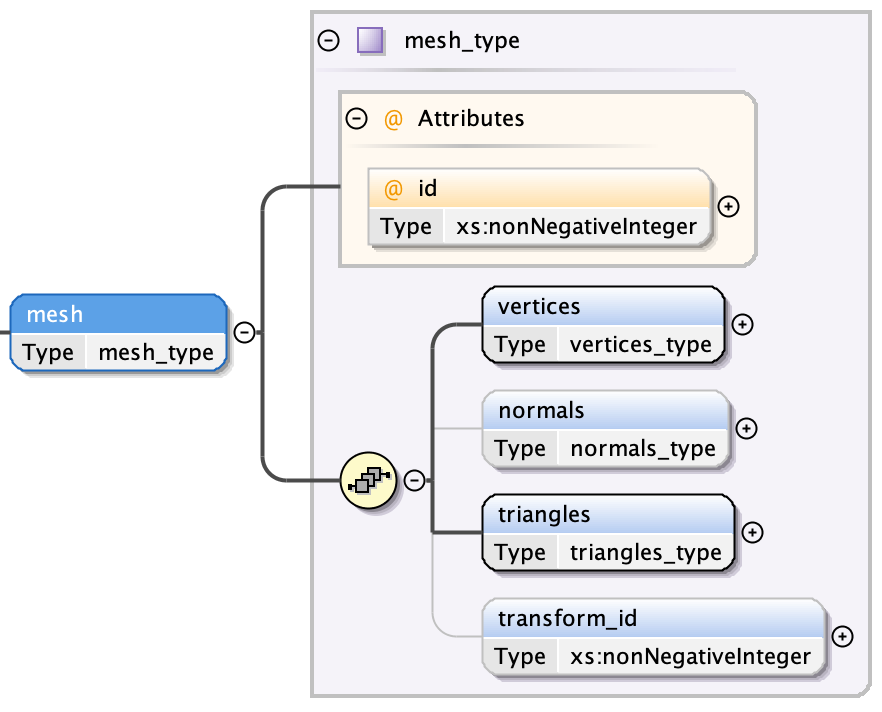
- a vertices attribute which holds an encoded list of float-triples (see Segments: Mesh Vertices)
- an optional corresponding normals attribute which holds an encoded list of float-triples (see Segments: Mesh Normals)
- a triangles attribue which contains an encoded list of int-triples (see Segments: Mesh Triangles)
- an optional transform index (from Transformation Matrix)
Segments: Mesh Vertices¶
A vertices object consists of five values:
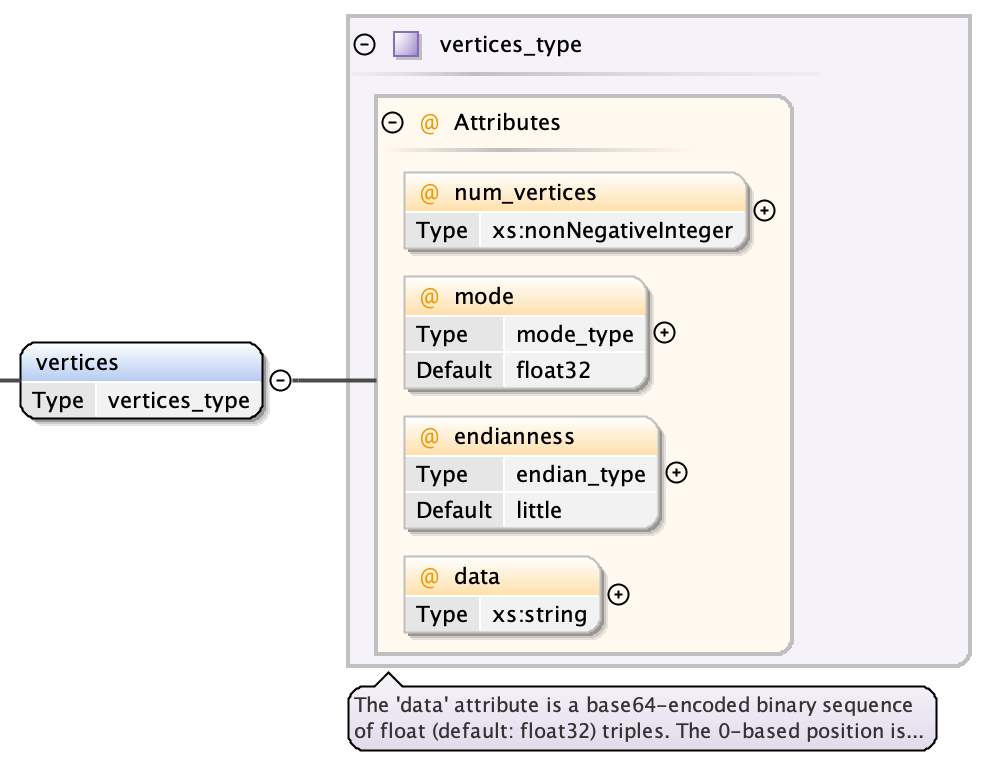
- a num_vertices attribute giving a count of the number of contained vertices
- a mode attribute (default “float32”) used for decoding the data
- a endianness attribute (default “little”) used for decoding the data
- the data as a base64-encoded binary string
Segments: Mesh Normals¶
An optional normals object exactly corresponds to the vertices object
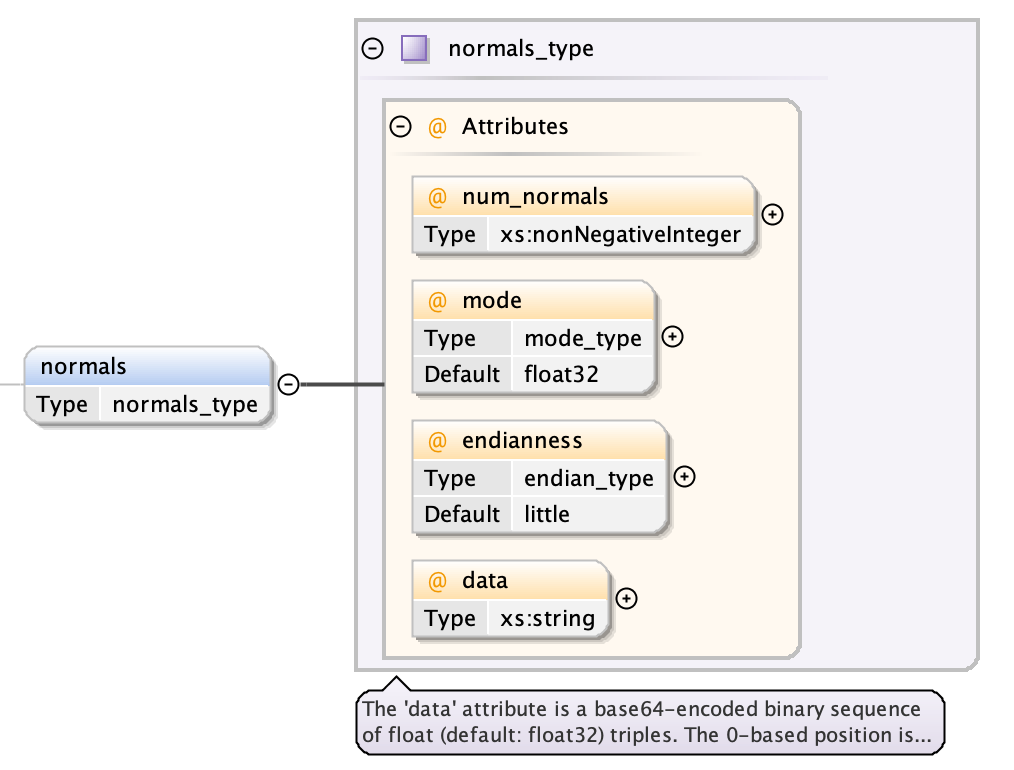
- a num_normals attribute giving a count of the number of contained normals
- a mode attribute (default “float32”) used for decoding the data
- a endianness attribute (default “little”) used for decoding the data
- the data as a base64-encoded binary string
Segments: Mesh Triangles¶
A triangle defines the topology of the points provided in the corresponding vertices object.
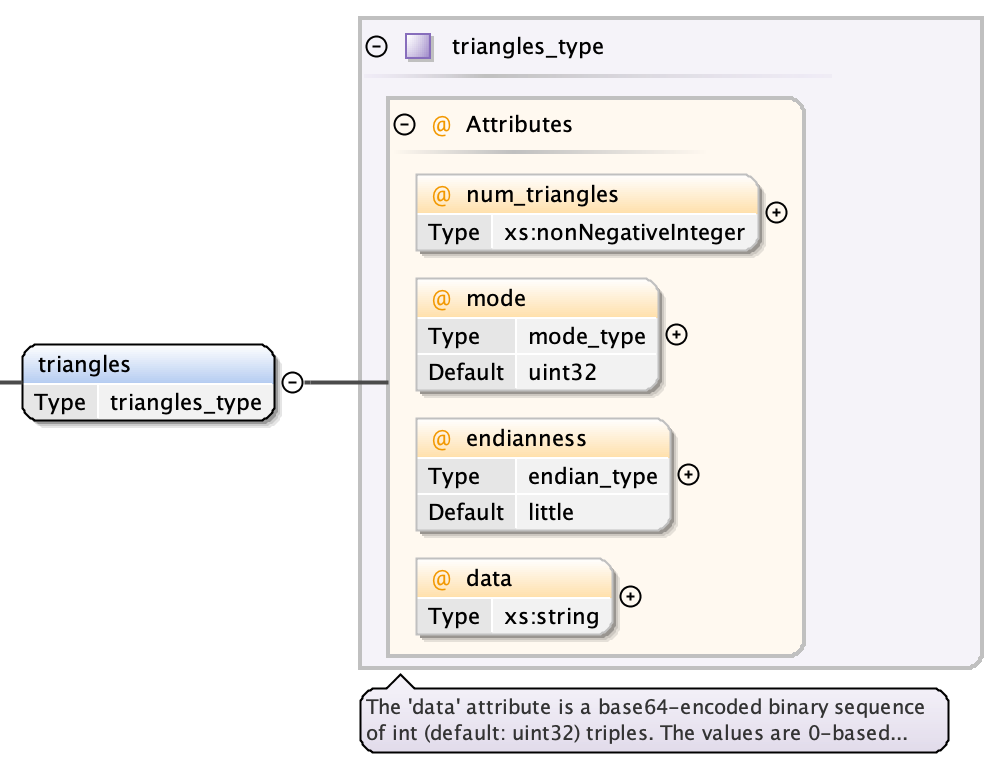
- a num_triangless attribute giving a count of the number of contained triangles
- a mode attribute (default “uint32”) used for decoding the data
- a endianness attribute (default “little”) used for decoding the data
- the data as a base64-encoded binary string
Segments: 3D Volumes¶
A 3D volume consists of three fields:
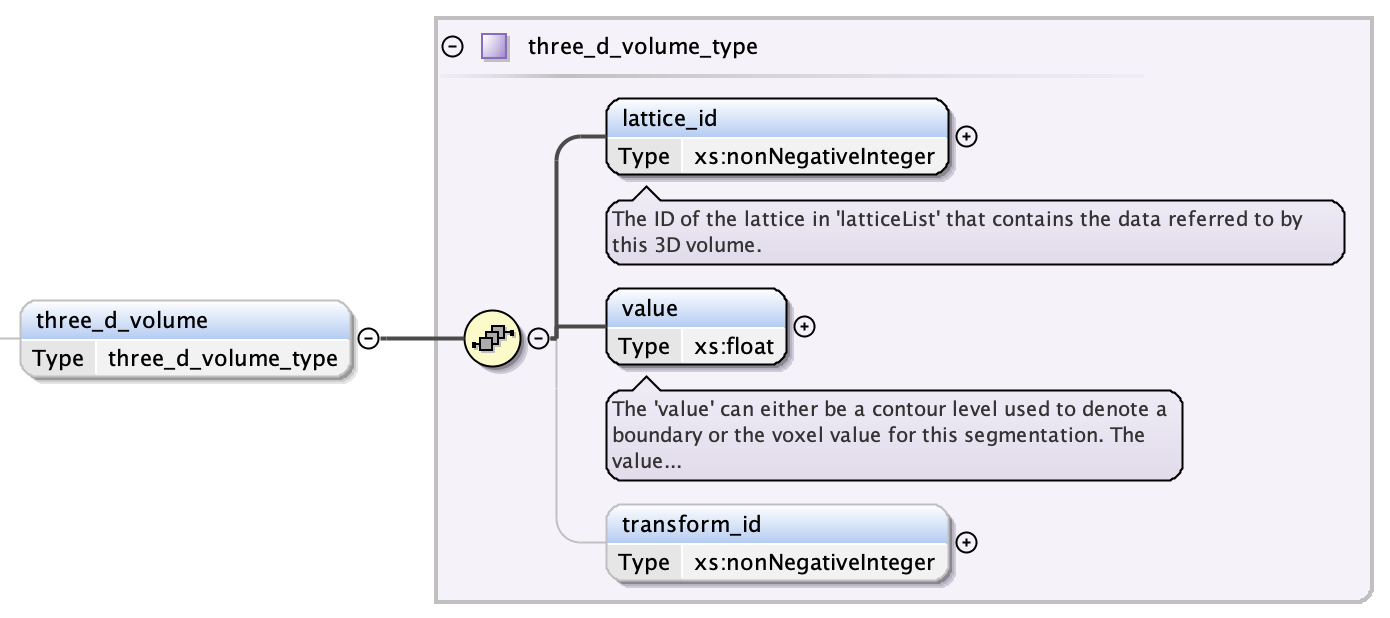
- the lattice index containing the volume data (see Lattices)
- the voxel value that identifies this 3D volume segment in the lattice specified in Lattices
- an optional transform index (from Transformation Matrix)
Lattices¶
A lattice describes a 3D structure consisting of orthogonally stacked voxels that has the following fields:
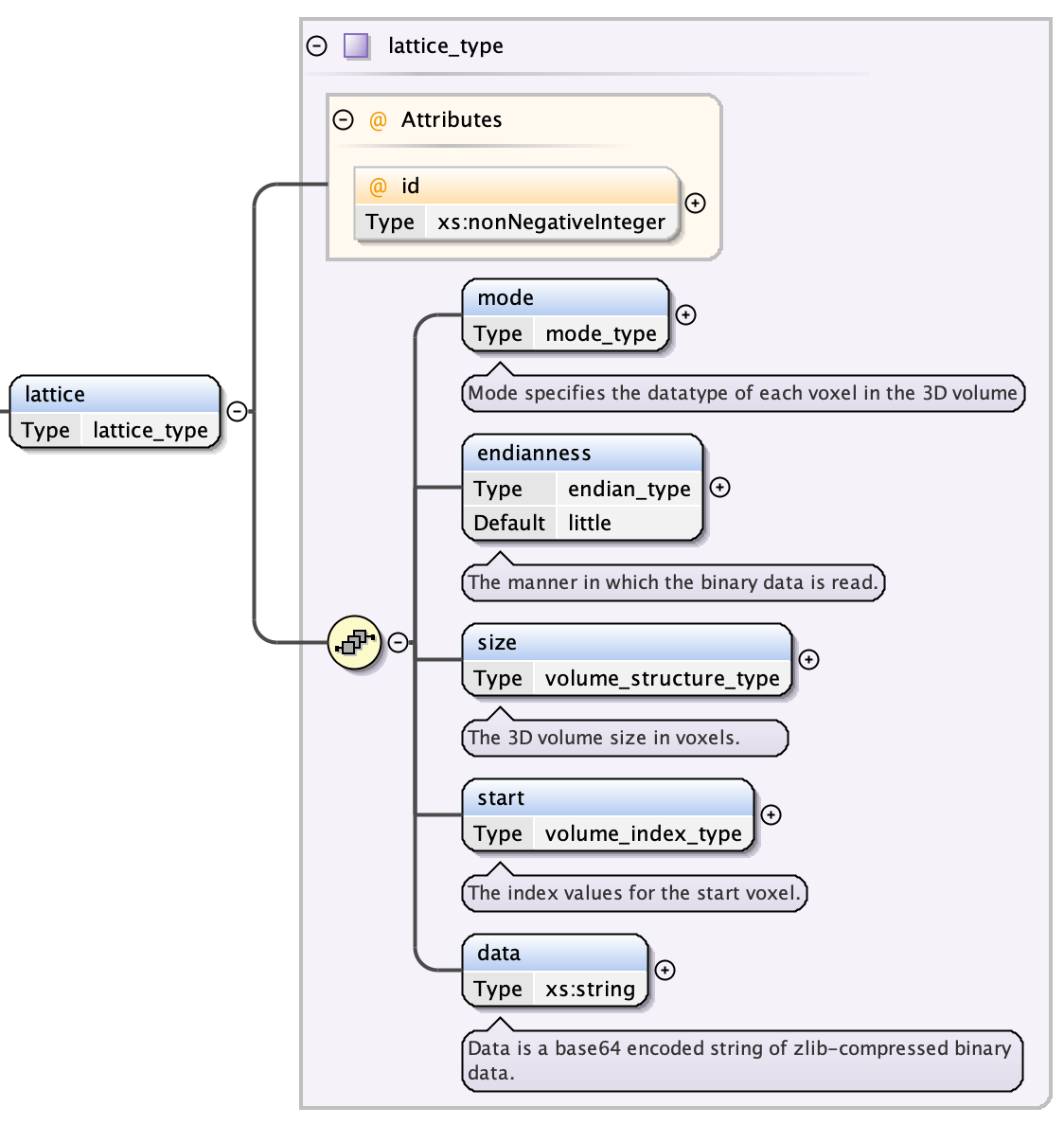
- a lattice index - a unique integer over all lattices
- a mode string which specifies the data type of each voxel; valid values are “int8”, “uint8”, “int16”, “uint16”, “int32”, “uint32”, “int64”, “uint64”, “float32”, “float64”
- the endianness of the lattice data; can be “little” or “big” (case-sensitive)
- the size of the lattice (see Lattice: Volume Size)
- the start indices of the lattice (see Lattice: Start Index)
- the data as a base64-encoded, zlib-zipped byte sequence
Lattice: Volume Size¶
The size has three fields:
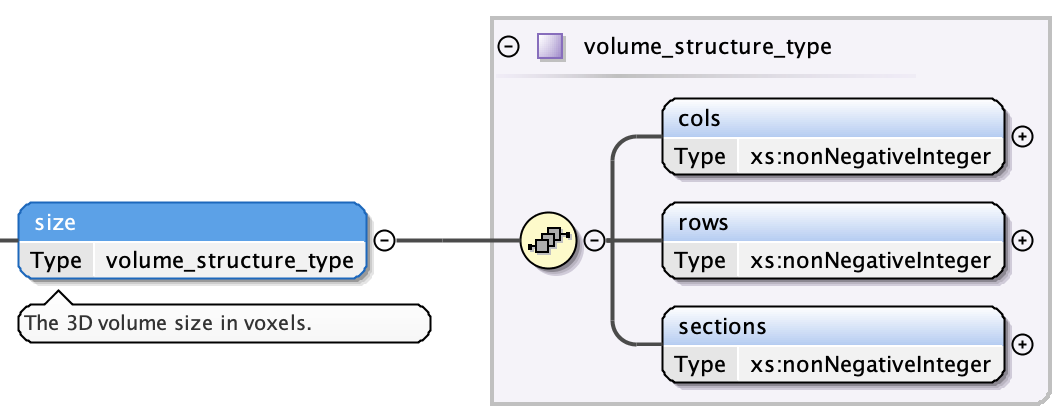
- the number of columns as a non-negative integer
- the number of rows as a non-negative integer
- the number of sections as a non-negative integer
Lattice: Start Index¶
The start indices have three fields:
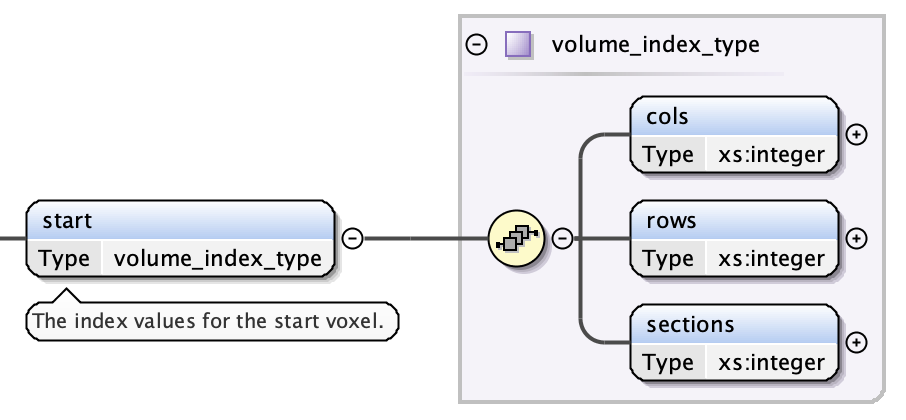
- the 0-based column index of the first voxel - an integer
- the 0-based row index of the first voxel - an integer
- the 0-based section index of the first voxel - an integer
Segments: Shape Primitives¶
There are different types of shape primitives. Each shape primitive has:
- an index - a unique integer value over all shape types
cones have:
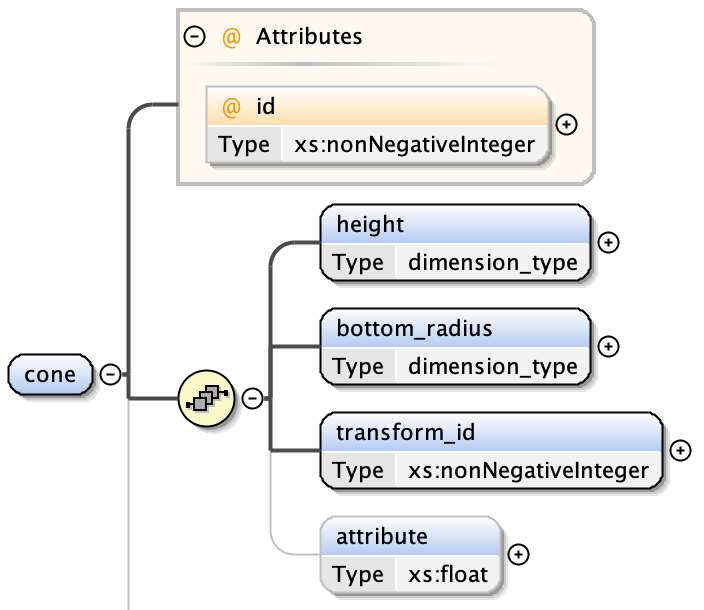
- a height
- a bottom radius
- an optional transform index (from Transformation Matrix)
- an optional attribute value - a floating point number associated with the shape
cuboids have:
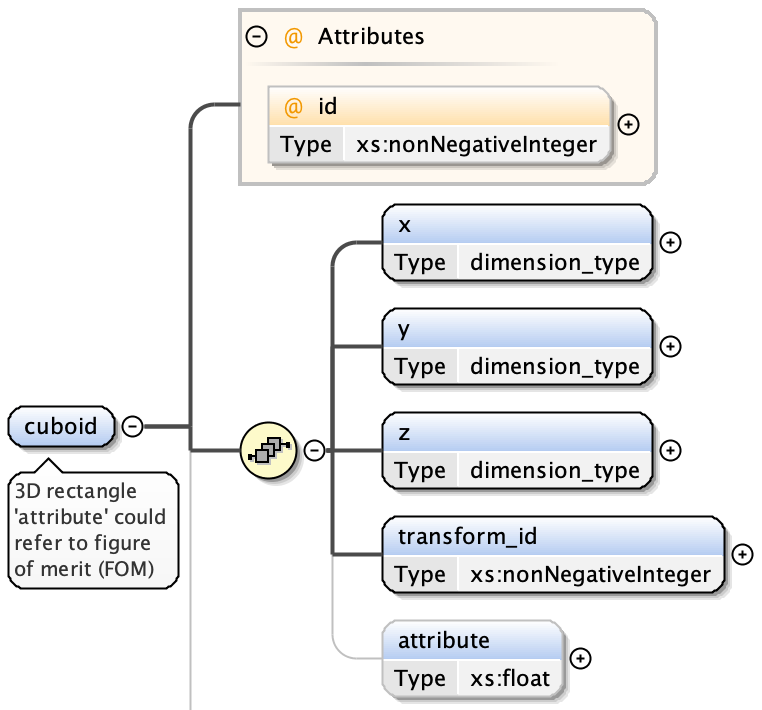
- x extent
- y extent
- z extent
- an optional transform index (from Transformation Matrix)
- an optional attribute value - a floating point number associated with the shape
cylinders have:
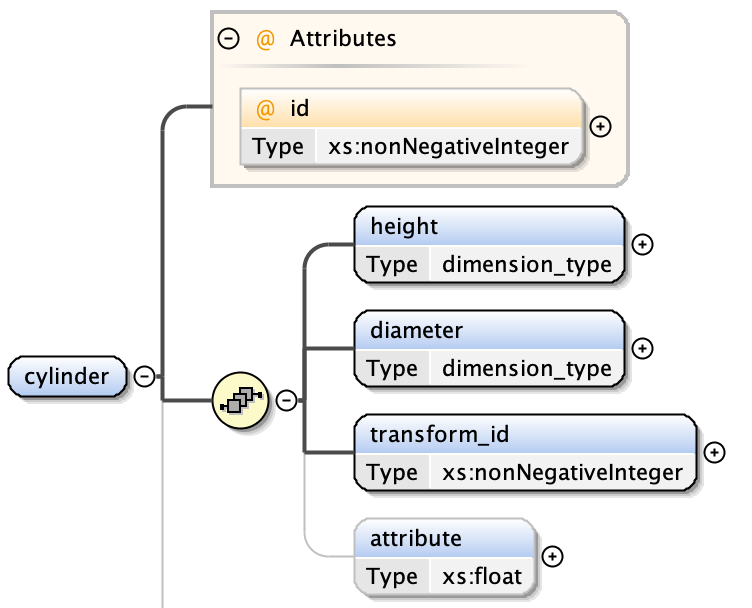
- a height
- a diameter
- an optional transform index (from Transformation Matrix)
- an optional attribute value - a floating point number associated with the shape
ellipsoids have:
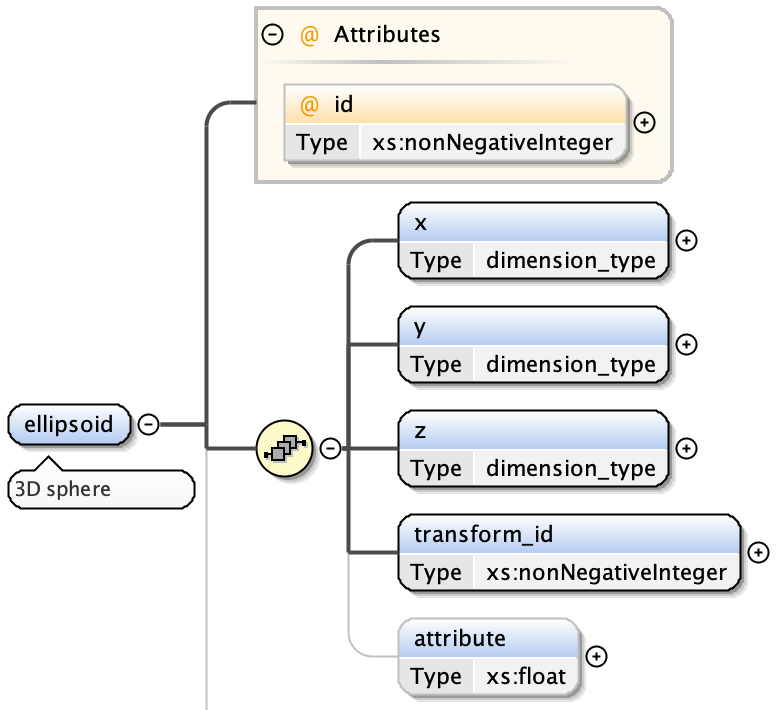
- x extent
- y extent
- z extent
- an optional transform index (from Transformation Matrix)
- an optional attribute value - a floating point number associated with the shape
subtomogram averages are exactly the same as 3D volumes and have:
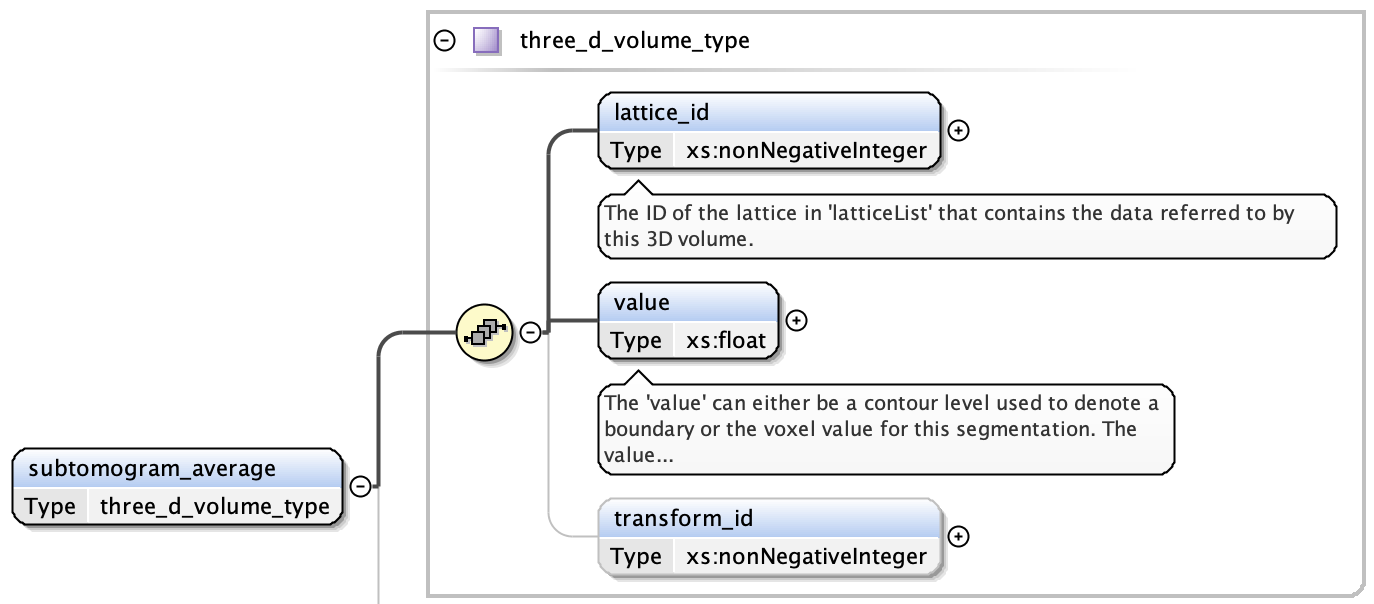
- the lattice index containing the volume data (see Lattices)
- the voxel value that specifies the recommended contour level in the lattice specified in Lattices
- an optional transform index (from Transformation Matrix)
Warning
This element has not been implemented in sfftk-rw.
Resources for External References¶
Here is a partial list of resources that may be used for (global) external references. Note that the main means by which these would be obtained is either using the sfftk notes utility or the online SAT (https://wwwdev.ebi.ac.uk/pdbe/emdb/sat_branch/sat/).
| Name | type | otherType | value | label | description |
|---|---|---|---|---|---|
| Ontologies available through EMBL-EBI’s Ontology Lookup Service (OLS) | <ontology_id> | http://purl.obolibrary.org/obo/<accession> | <accession> | label field |
description field |
| Gene Ontology (GO) | go | http://purl.obolibrary.org/obo/<GO_accession> | <GO_accession> | label field |
description field |
| Electron Microscopy DataBank (EMDB) | emdb | https://www.ebi.ac.uk/pdbe/entry/emdb/<EMDB_accession> | <EMDB_accession> | search_term |
title field |
| UniProt | uniprot | https://www.uniprot.org/uniprot/<UniProt_accession> | <UniProt_accession> | name field |
proteins field |
| Protein Data Bank | pdb | https://www.ebi.ac.uk/pdbe/entry/pdb/<PDB_accession> | <PDB_accession> | search_term |
title field |
| Europe PMC | europepmc | https://europepmc.org/abstract/MED/<EuropePMC_accession> | <EuropePMC_accession> | authorString field |
title field |
| Electron Microscopy Public Image ARchive (EMPIAR) | empiar | https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/<EMPIAR_accession> | <EMPIAR_accession> | search_term |
title field |